

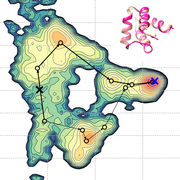
We identified "refinement pathways" that connect the native states and the initial models for 8 protein structure models. Surprisingly, but we expected, the native states have the lowest free energy with the latest CHARMM force field (c36m) for all the tested target. The corresponding structures have sub-Angstrom similarity to the experimental structures in many of the cases. Moreover, we figured out that protein structures should be partially unfolded and refolded to get to the native state along the refinement pathways. And, it takes a few to hundreds of microsecond depending on the protein size and complexity of modeling errors. Read the full story here.